A molecular dynamics framework to explore the structure
and dynamics of layered double hydroxides
and dynamics of layered double hydroxides
by Germán Pérez-Sánchez, Tiago L. P. Galvão, João Tedim, José R. B. Gomes
News
The window for applications for a new post-doctoral research grant to perform classical MD simulations within the framework of project SELMA - Unveiling the self-healing mechanisms associated with smart nanocontainers, PTDC/QEQ-QFI/4719/2014, financed by national funds through FCT/MEC (PIDDAC) and, when applicable, co-financed by FEDER under the PT2020 Partnership Agreement, is from January 17th to January 31th. Please follow this link for additional details.Last Articles
from project SELMAPhys. Chem. Chem. Phys., 19 (2017) 6113.
J. Phys. Chem. C, 121 (2017) 2211.
Theor. Chem. Acc., 135 (2016) 78.
LDH models: Input files for MD simulations using Gromacs
For each system, the package includes the topology along with an equilibrated simulation box for running MD simulations with the Gromacs code (.top, .itp and .gro files). A script (.sh file) to run equilibration and production runs (.mdp files) is also provided. The Gromacs version used in the calculations was the 5.1.4. and, very probably, problems may appear if the packages are used with previous versions of the code.
Table of Contents:
- i) Cell parameters (*.gro): The initial LDH supercell is readytominimization.gro and the structure obtained after the MD simulations is md.gro.
- ii) Molecular dynamics parameters input files (*.mpd):
- iii) Input topology parameter files: The force field parameters for the LDH can be found in LDH.ff folder and the partial charges in the LDH.itp file.
- iv) Run the MD simulations: equilib_production.sh - Script (bash shell) to compile and run the MD simulations.
- ii.i) Energy minimization:
em1.mdp - The metal atoms and the hydroxide (OH) groups are constrained at their DFT optimized positions and only the water molecules and the ions are allowed to move.
em2.mdp - Only the metal atoms are frozen and the remaining atoms are allowed to move.
ii.ii) NVT equilibration:
nvt1.mdp - The metal atoms and the OH groups are frozen while freely moving the remaining components of the system (1 ns).
nvt2.mdp - Only the positions of the metal atoms are constrained (2.5 ns).
ii.iii) NpT equilibration:
npt.mdp - 50 ns of MD simulation for the density equilibration.
ii.iv) NpT production:
md.mdp - 50 ns of MD simulation for production.
Parameters for anions can be found in cl.itp, nitrate.itp and CO3.itp for chloride, nitrate and carbonate anions, respectively.
The water model is included in oplsaa.ff folder.
The file topol.top includes the paths to the files and folders with the potential parameters together with the number of components in the simulation box used in the MD simulations.
For those just starting with molecular dynamics simulations with the Gromacs code it is highly recommended to have a look on the information provided in the following webpage.
| System | Supercell | Notation | Package |
|---|---|---|---|
| Mg2Al-Cl | [Mg4Al2(OH)12]Cl2.4H2O | MI | MI.zip |
| Mg2Al-Cl | [Mg4Al2(OH)12]Cl2.2H2O | MI1/2 | MI1_2.zip |
| Mg2Al-NO3 | [Mg4Al2(OH)12](NO3)2.4H2O | MII | MII.zip |
| Mg2Al-CO3 | [Mg4Al2(OH)12]CO3.4H2O | MIII | MIII.zip |
| Zn2Al-Cl | [Zn4Al2(OH)12]Cl2.4H2O | ZI | ZI.zip |
| Zn2Al-Cl | [Zn4Al2(OH)12]Cl2.2H2O | ZI1/2 | ZI1_2.zip |
| Zn2Al-NO3 | [Zn4Al2(OH)12](NO3)2.4H2O | ZII | ZII.zip |
| Zn2Al-CO3 | [Zn4Al2(OH)12]CO3.4H2O | ZIII | ZIII.zip |

![]() 21.10.2017
21.10.2017
Views of the MI-MIII supercells used in molecular dynamics simulations. These supercells were obtained from the primitive cells (*.pdb) optimized with the Quantum Espresso code using the GGA-PBE density functional.
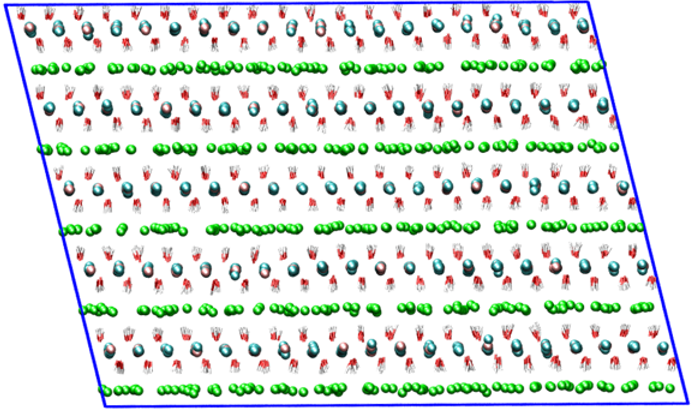 | 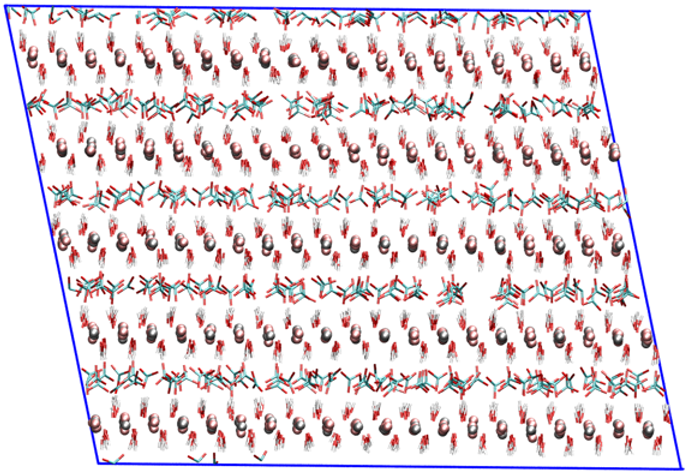 | 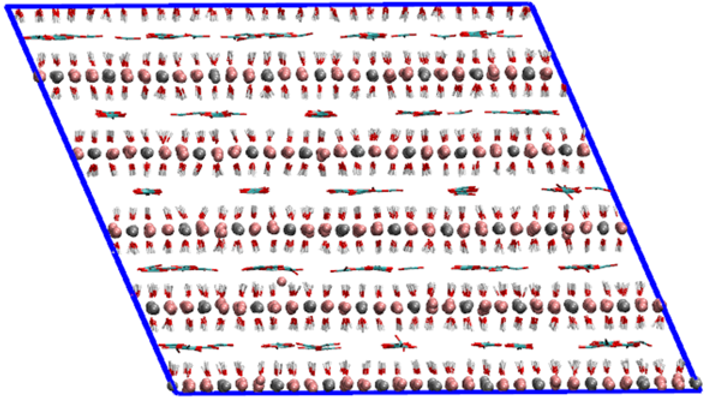 |
| MI | MII | MIII |

![]() 21.10.2017
21.10.2017